Standardised survival probabilities in R
Hi everyone,
You might already know that a couple of years ago Betty Syriopoulou published a tutorial paper on standardised survival probabilities, a useful tool to supplement and enhance the reporting of regression models for time-to-event (survival) outcomes.
The background, in short, is that while hazard ratios are ubiquitous in medical research with time-to-event outcomes, they are often misreported, misinterpreted, and have several, well-known limitations; I will not go much into detail, but some relevant references if you want to read more about this topic are:
- Betty’s tutorial paper on standardised survival probabilities (of course);
- The popular paper by Miguel Hernán on The Hazards of Hazard Ratios;
- The paper by Mats Stensrud and Miguel Hernán questioning Why Test for Proportional Hazards.
Standardised survival probabilities provide a useful and easy-to-interpret measure to complement hazard ratios, can be calculated after fitting survival models (and can therefore include covariates to adjust for, e.g., because they are confounders for a certain exposure-outcome association of interest), and can be contrasted to produce standardised survival probability differences (i.e., risk differences) and ratios (i.e., risk ratios). As a bonus, if the confounders that you decide to include in your regression model are sufficient to control for confounding, then the estimated risk differences (or ratios) can be interpreted as population causal effects: nice! More on this causal interpretation here and here.
The tutorial paper is easy to follow and illustrates the utility of such measures very nicely, but most importantly, Stata code to reproduce the analyses and use this method in practice is provided in the supplementary material.
Unfortunately, only Stata code was published with the paper, but Betty and I have been talking about porting this to R for a while now, and guess what: today is your lucky day! We have just published on my GitHub profile a repository with R code that can be used to replicate every result from the paper, including every plot that was included in the tutorial. The GitHub repository can be found here (https://github.com/ellessenne/standsurv-tutorial-r), and the code is released under the MIT license, so you can pretty much do whatever you want with it.
Here is a little preview of what you will be able to accomplish:
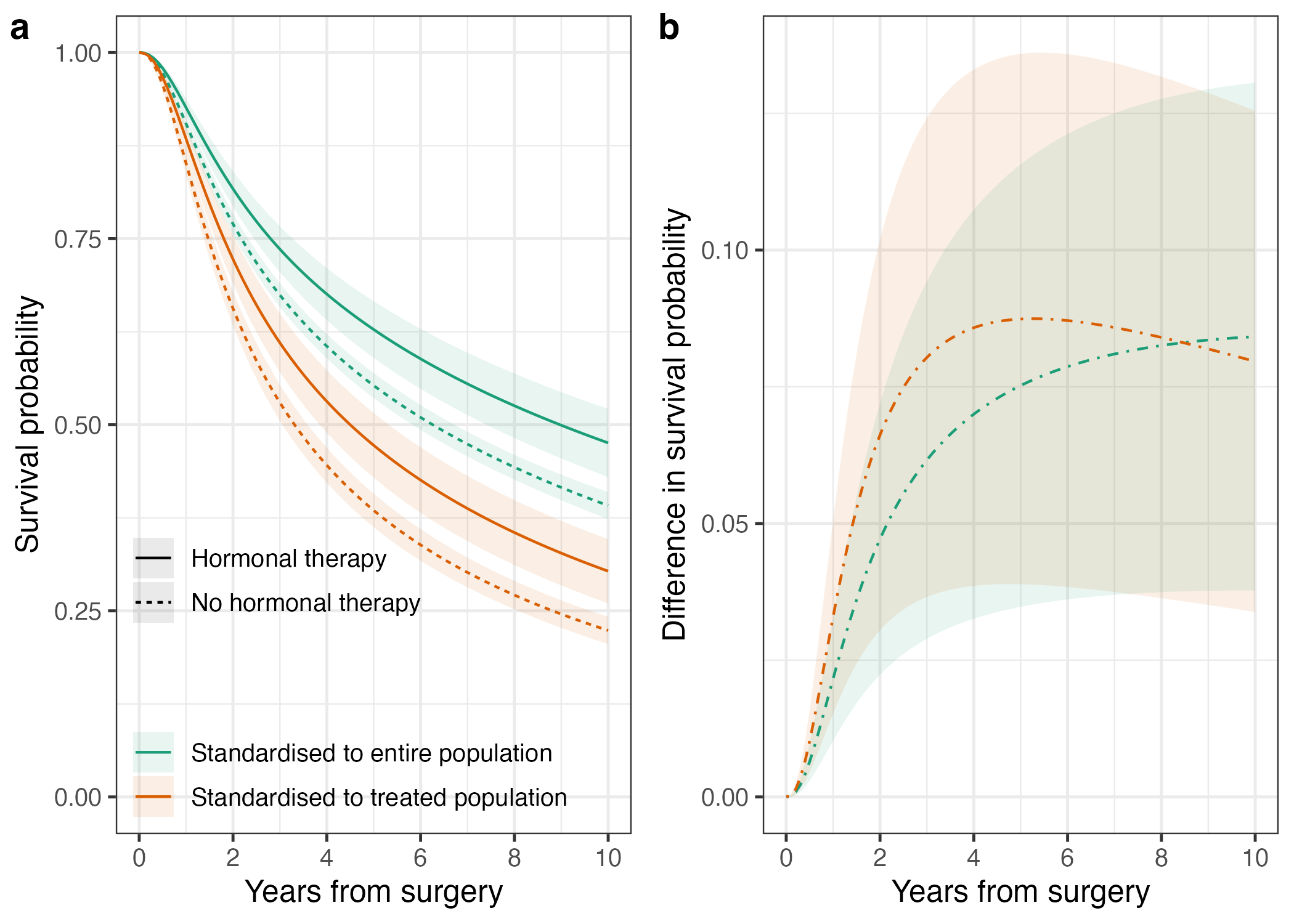
…and there you have it, don’t forget to check out the code and go test this out for your next project, predict standardised probabilities, make some nice plots, complement your hazard ratios, and take your survival analyses to the next level.
Finally, please note that the code in the repository is quite specific to this project and not very general, so you will have to do a little bit of work to get this going, but there should be enough comments to give you all the information you will possibly need to extend it. If you have any questions, feel free to get in touch or just open an issue in the GitHub repository. As always, any feedback is more than welcome.
Before we go, here is one more treat for you, if you are still reading: an R package with a general, easy to use implementation of regression standardisation may or may not be in development. Stay tuned for more… 👀